OCPID Guide
What is OCPID?
Organ Centric Protein Interaction Database (OCPID) is a bioinformatics web tool, developed to provide integrated analysis of human protein-protein interaction data and organ/tissue specific gene expression datasets.
- Integrated human protein databases: DIP, HPRD, MINT, INTACT
- Organ/tissue specific gene expression datasets:
- Shyamsundar et al., Genome Biol, 2005;6:R22.
- Son et al., Genome Res., March 1, 2005; 15(3):443-450.
- Su et al., Proc. Natl. Acad. Sci. USA 101 (2004), 6062-6067
Quick Start
Select a protein ID type, enter an ID into the value field on the OCPID Home/New Search page, check Organs, and click the Submit button in order to begin the search.
The Search Result page displays a protein-protein interaction list of all genes matching your query. The OCPID displays some links and buttons such as average gene expression charts, protein interaction only list, and protein interaction network graph.
Click the links and buttons you wish to explore further.
Search
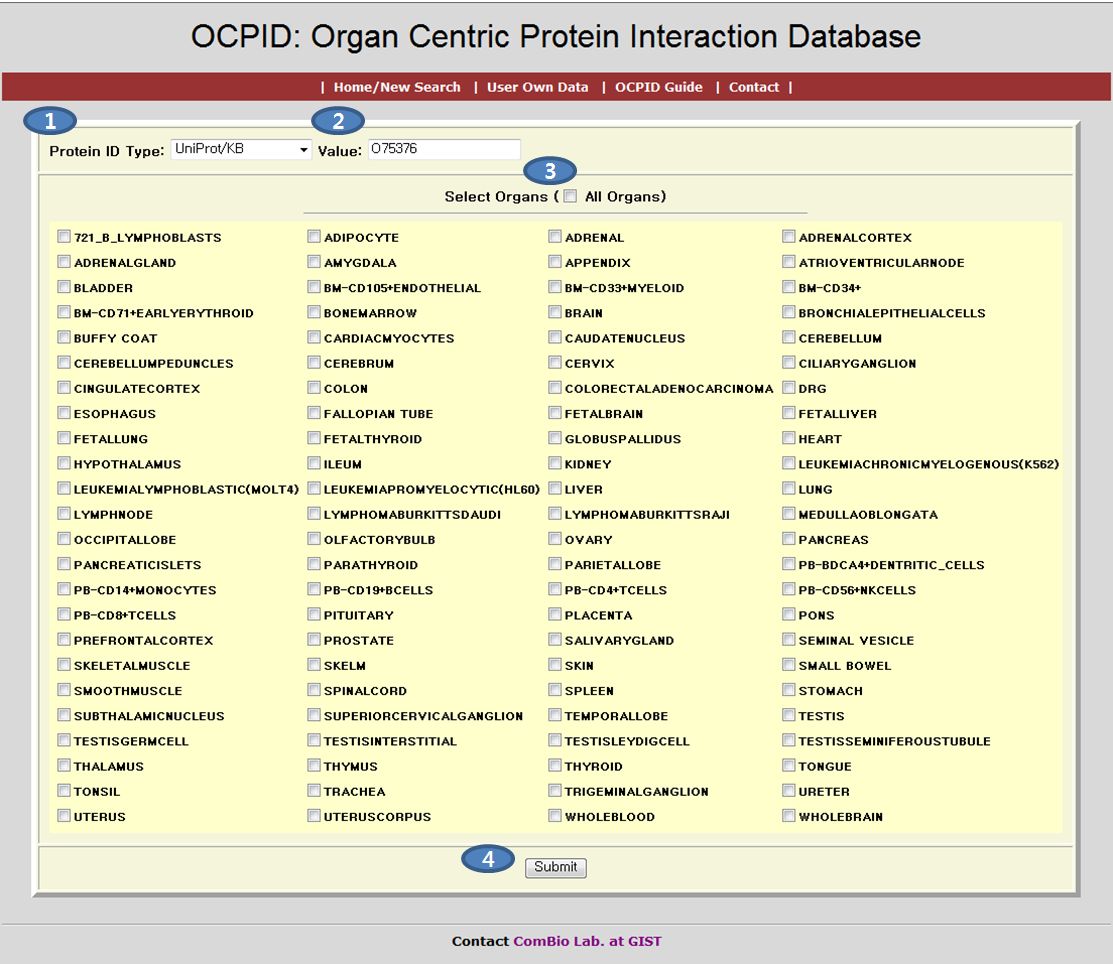
On the OCPID Home/New Search page, you can get organ specific protein interaction list by following simple steps.Choose a protein ID type in the combobox item. It has four types of protein IDs (Unigene, Gene Symbol, Uniprot, and Refseq).
Enter an ID into the value field. You should input a correct ID that is matched with the protein ID type that you choose.
Select check Organs on the check box by the organ name. If you want to search all organs, you can just select a check box by 'All organs' under the value field.
Click the Submit button to start the search.
What do I get?
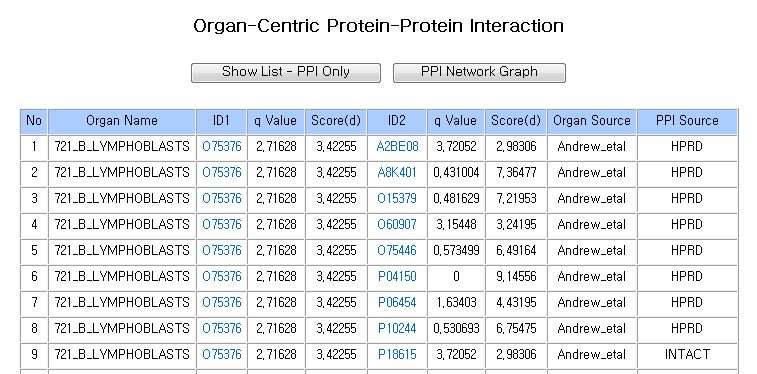
Basically, you can have four types of search results.The list of Organ-Centric Protein-Protein Interaction is displayed on the search result page. It contains valuable informations such as organ specific protein interactions, significant values and distance score by SAMR. It also display the source of Gene Expression Datasets and Protein Interaction Databases.
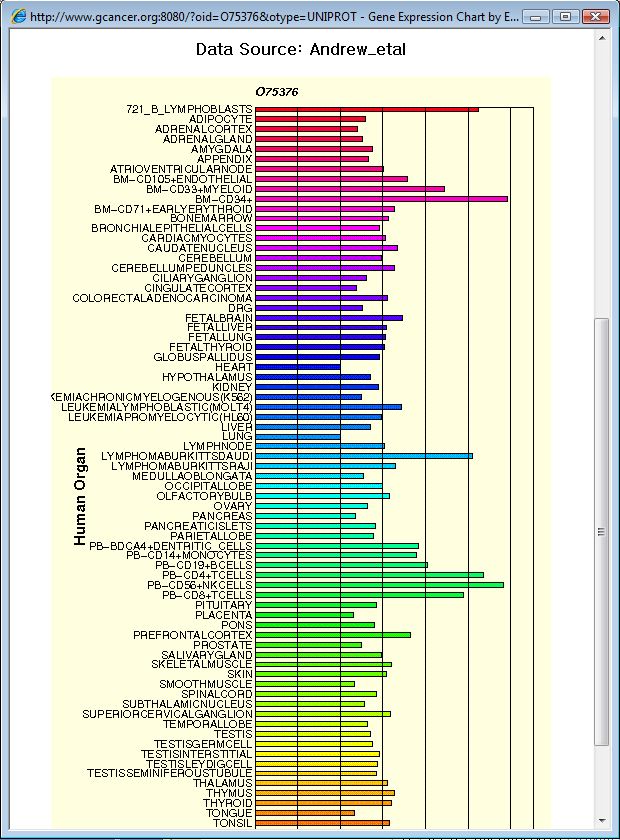
The chart for average gene expression by each organ can be displayed by clicking the link at the protein ID on the list table.
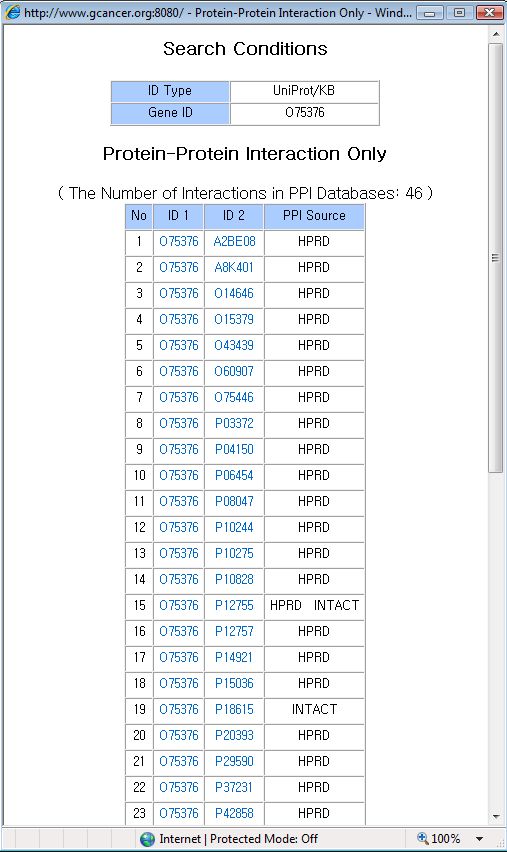
The list of ppi only can be displayed by click the 'Show List - Ppi Only' button. It could be used when you want to search all protein interactions which is not related with ogran specific information.
The protein network graph can be displayed by clicking the 'PPI Network Graph' button. It requires the java runtime environments(JRE). If you don't have it, the displayed page will ask downloading and installing JRE programs. Sometimes, you need to check and allow your browser's security options.
|
|
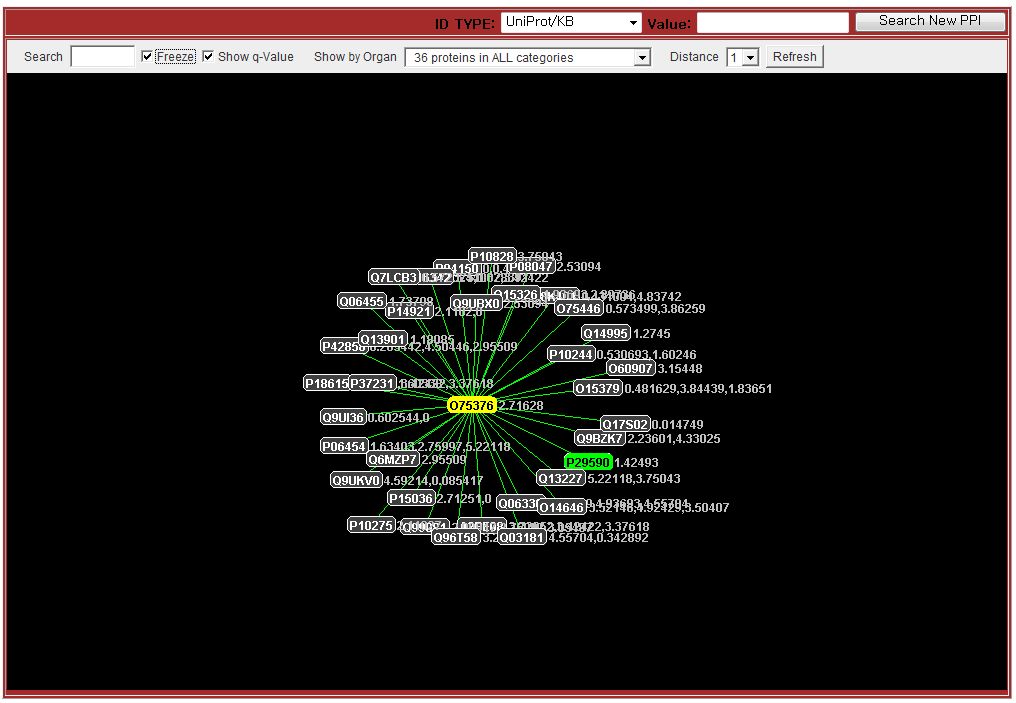
Search Examples
You can test OCPID by following sample IDs to have more information.Unigene: Hs.170839, Hs.289271, Hs.171995
Gene Symbol: E2F1, NHF1, EGFR, ANK1
Uniprot: O75376, P00742, P00734, P04150
Refseq: NP_005905.2, NP_00497.1, NP_000030.1
How to Registrate?
You can make your ID easily. You just click the 'User Own Data' item in the top menu bar, and click the '[New Registration]'. It will display a registration page for you.
Login to OCPID
If you have a ID on OCPID, you can login with your email ID and password. OCPID will show the data list of your own datasets which are uploaded before.
Search with your dataset
If you have data list, you can search protein interactions that are related with your data. It is same as OCPID's 'home/new search' page.
Add your dataset
If you login with your ID, you can upload and analyze your dataset by click the 'Add Data' menu. it requires more informations for correct analysis as shown in the following figure.
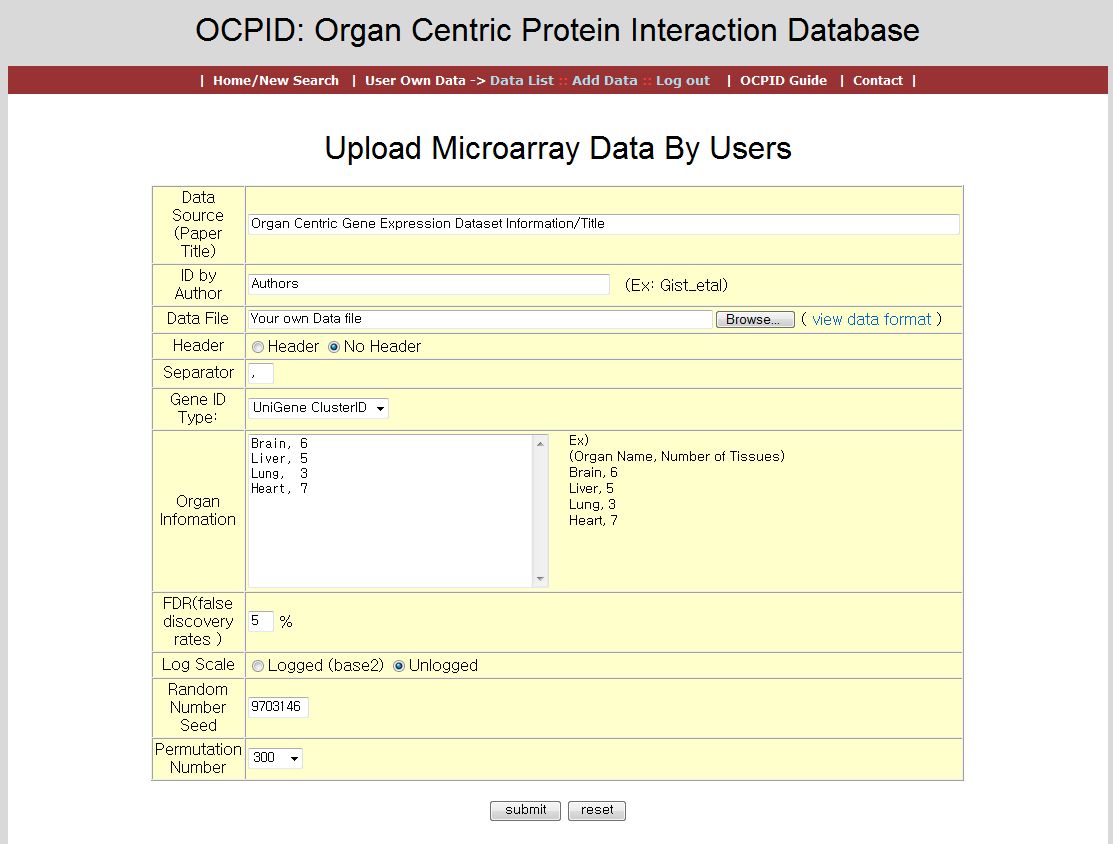
If all the process is done, it shows the messages about analysis and the information of database works. If it is done well, you can search organ centric protein interaction with your new dataset.
Examples to Test User Dataset
You can test OCPID by following sample datasets to have more information. You can download by mouse right button click. If you use these sample dataset (permutation number: 100), it may take about 15~25 minute to analysis to find significant genes.Unigene: Unigene Cluster Dataset(2.57MB)
Gene Symbol: Gene Symbol Dataset(2.51MB)
Uniprot: Uniprot Dataset(2.54MB)
Refseq: Refseq Dataset(2.57MB)